DNA Binding Motif
| Accessions: | GCM2_DLX3_1 (HumanTF2 1.0) |
| Names: | GCM2_DLX3 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 19 |
| Consensus: | aTrcGGGywkwyttAATtr |
| Weblogo: | 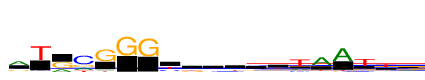 |
| PSSM: | P0 A C G T 01 2925 671 1195 163 a 02 75 305 133 4120 T 03 1338 206 2782 0 r 04 1320 4120 1341 836 c 05 437 36 4120 361 G 06 0 24 4120 6 G 07 0 26 4120 59 G 08 621 1275 347 1877 y 09 1271 904 825 1120 w 10 968 812 1088 1253 k 11 1283 766 875 1196 w 12 893 1114 1019 1094 y 13 925 709 1008 1479 t 14 1109 763 359 4120 t 15 4120 354 647 586 A 16 4120 32 232 429 A 17 511 467 793 4120 T 18 844 534 937 3183 t 19 1959 437 2161 861 r |
| Type: | Heterodimer |
| Binding TFs: | DLX3_TF2 (Homeobox domain) GCM2_TF1 (GCM motif protein) DLX3_Crystal (Homeobox domain) DLX3_Full (Homeobox domain, Homeobox protein distal-less-like N terminal ) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.