Main news:
19/08/2025 Updated 3d-footprint & interface residues
29/07/2025 Added REST API supporting protein queries
22/07/2025 Updated EEADannot with plant motifs and sites manually curated at EEAD-CSIC
07/01/2025 synced with RSAT (see blog), previous contents archived at DIGITAL.CSIC
05/01/2024 Added JASPAR 2024 (see blog)
..
09/10/2023 Adopted python3-weblogo & ghostscript
31/05/2023 Updated motif & FASTA downloads after reviewing licenses
03/03/2022 Updated user manual, see how to build a URL for a motif at our blog
19/08/2025 Updated 3d-footprint & interface residues
29/07/2025 Added REST API supporting protein queries
22/07/2025 Updated EEADannot with plant motifs and sites manually curated at EEAD-CSIC
07/01/2025 synced with RSAT (see blog), previous contents archived at DIGITAL.CSIC
05/01/2024 Added JASPAR 2024 (see blog)
..
09/10/2023 Adopted python3-weblogo & ghostscript
31/05/2023 Updated motif & FASTA downloads after reviewing licenses
03/03/2022 Updated user manual, see how to build a URL for a motif at our blog
The data are regularly curated from public databases and the literature; interfaces are inferred from the collection of protein-DNA complexes at 3D-footprint:
| total | unique | metazoa | plants | |
|---|---|---|---|---|
| Transcription Factors | 10168 | 7503 | 4839 | 1437 |
| DNA motifs (PSSM) | 17506 | 14828 | 8893 | 3346 |
| DNA Binding Sites/Sequences | 68358 |
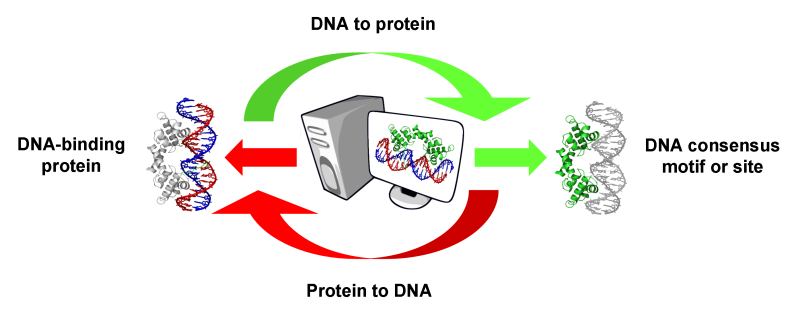
footprintDB predicts:
- Transcription factors which bind a specific DNA site or motif
- DNA motifs or sites likely to be recognized by a specific DNA-binding protein
 |
||
|
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.

