DNA Binding Motif
| Accessions: | TFAP2C_ONECUT2_2_3 (HumanTF2 1.0) |
| Names: | TFAP2C_ONECUT2 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 26 |
| Consensus: | tATCGAyyggwyrtysCCysrGGsga |
| Weblogo: | 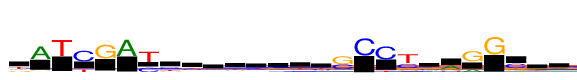 |
| PSSM: | P0 A C G T 01 292 221 183 619 t 02 1315 0 115 88 A 03 55 9 0 1315 T 04 34 1315 8 372 C 05 90 18 1315 32 G 06 1315 0 0 56 A 07 33 613 0 1315 y 08 186 583 177 368 y 09 237 298 556 224 g 10 315 301 405 294 g 11 411 179 320 406 w 12 231 389 267 428 y 13 371 214 467 263 r 14 273 196 238 608 t 15 302 400 204 408 y 16 191 821 1315 95 s 17 8 1315 13 0 C 18 8 1315 17 181 C 19 114 655 494 1315 y 20 310 334 477 194 s 21 732 206 333 44 r 22 392 34 1315 15 G 23 13 0 1315 0 G 24 31 802 513 79 s 25 314 286 401 314 g 26 481 292 232 311 a |
| Type: | Heterodimer |
| Binding TFs: | ONECUT2_TF2 (Homeobox domain, CUT domain) TFAP2C_TF1 (Transcription factor AP-2) ONECUT2_Full (Homeobox domain, CUT domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.