DNA Binding Motif
| Accessions: | TFAP2C_DLX3_1 (HumanTF2 1.0) |
| Names: | TFAP2C_DLX3 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 24 |
| Consensus: | yssCysvrGGCaygggssTAATkr |
| Weblogo: | 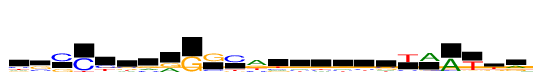 |
| PSSM: | P0 A C G T 01 241 341 219 315 y 02 173 407 709 93 s 03 50 1116 474 7 s 04 4 1116 3 138 C 05 61 503 156 395 y 06 238 313 292 273 s 07 330 315 382 89 v 08 550 26 566 29 r 09 3 10 1116 0 G 10 78 296 1116 31 G 11 9 1116 193 280 C 12 1116 335 601 477 a 13 264 299 147 406 y 14 270 252 354 239 g 15 199 239 422 255 g 16 275 240 366 234 g 17 195 284 408 229 s 18 267 310 303 236 s 19 85 203 74 1116 T 20 1116 100 143 85 A 21 1116 0 40 76 A 22 109 53 130 1116 T 23 150 203 412 704 k 24 576 120 539 87 r |
| Type: | Heterodimer |
| Binding TFs: | DLX3_TF2 (Homeobox domain) TFAP2C_TF1 (Transcription factor AP-2) DLX3_Crystal (Homeobox domain) DLX3_Full (Homeobox domain, Homeobox protein distal-less-like N terminal ) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.