DNA Binding Motif
| Accessions: | GCM1_CEBPB_2_3 (HumanTF2 1.0) |
| Names: | GCM1_CEBPB |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 21 |
| Consensus: | rTRcGGGyrgkrTTGCGYAAy |
| Weblogo: | 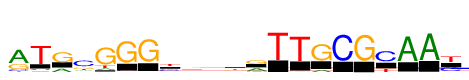 |
| PSSM: | P0 A C G T 01 1999 325 831 169 r 02 169 176 67 2323 T 03 620 58 1704 2 R 04 324 1738 586 494 c 05 281 52 2323 405 G 06 5 131 2323 84 G 07 34 70 2323 46 G 08 208 944 164 1008 y 09 775 346 862 340 r 10 536 500 781 507 g 11 313 254 838 919 k 12 892 275 1158 0 r 13 0 0 0 2323 T 14 5 3 102 2323 T 15 322 3 2001 4 G 16 0 2323 7 24 C 17 20 4 2323 8 G 18 56 1700 0 623 Y 19 2323 93 0 3 A 20 2323 10 2 4 A 21 5 898 122 1299 y |
| Type: | Heterodimer |
| Binding TFs: | CEBPB_TF2 (bZIP transcription factor, Basic region leucine zipper) GCM1_TF1 (GCM motif protein) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.