DNA Binding Motif
| Accessions: | ZNF740_4 (HT-SELEX2 May2017), MA0753.2 (JASPAR 2024) |
| Names: | ZNF740 |
| Organisms: | Homo sapiens |
| Libraries: | HT-SELEX2 May2017 1, JASPAR 2024 2 1 Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schübeler D, Vinson C, Taipale J. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science : (2017). [Pubmed] 2 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | HT-SELEX |
| Length: | 13 |
| Consensus: | cyrCCCCCCCCAc |
| Weblogo: | 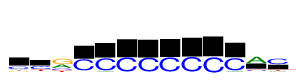 |
| PSSM: | P0 A C G T 01 186 502 182 185 c 02 162 677 72 378 y 03 727 343 1055 312 r 04 60 1055 37 49 C 05 110 1055 14 1 C 06 18 1055 4 16 C 07 22 1055 7 15 C 08 23 1055 3 11 C 09 10 1055 7 6 C 10 2 1055 3 3 C 11 115 1055 6 4 C 12 1055 239 68 134 A 13 207 1055 54 246 c |
| Type: | Heterodimer |
| Binding TFs: | ZNF740 (Zinc finger, C2H2 type, Zinc-finger double domain, C2H2-type zinc finger) Q8NDX6 (Zinc finger, C2H2 type, Zinc-finger double domain, C2H2-type zinc finger) Q8NDX6 |
| Publications: | Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schübeler D, Vinson C, Taipale J. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science : (2017). [Pubmed] Weirauch MT, Cote A, Norel R, Annala M, Zhao Y, Riley TR, Saez-Rodriguez J, Cokelaer T, Vedenko A, Talukder S; DREAM5 Consortium, Bussemaker HJ, Morris QD, Bulyk ML, Stolovitzky G, Hughes TR. Evaluation of methods for modeling transcription factor sequence specificity. Nat Biotechnol 31:126-34 (2013). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.