DNA Binding Motif
| Accessions: | TFAP2C_ELK1 (HumanTF2 1.0) |
| Names: | TFAP2C_ELK1 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 20 |
| Consensus: | yGCCksrGGsgrCGGAAGyg |
| Weblogo: | 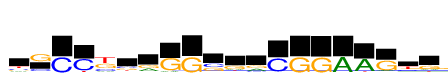 |
| PSSM: | P0 A C G T 01 3302 4487 3570 6320 y 02 890 6236 17679 608 G 03 0 17679 42 0 C 04 39 17679 178 2593 C 05 3315 7010 13653 17679 k 06 2678 5960 5783 3258 s 07 6878 1950 8160 691 r 08 2097 4 17679 0 G 09 0 0 17679 24 G 10 639 8977 8702 897 s 11 3693 1792 9536 2658 g 12 6642 3064 7434 539 r 13 874 17679 241 108 C 14 5 22 17679 14 G 15 9 54 17679 48 G 16 17679 0 0 112 A 17 17679 327 55 1515 A 18 2801 1060 17679 139 G 19 1500 5812 3662 11867 y 20 3708 1503 9405 3064 g |
| Type: | Heterodimer |
| Binding TFs: | ELK1_TF2 (Ets-domain) ELK1_Full (Ets-domain) TFAP2C_TF1 (Transcription factor AP-2) ELK1_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.