DNA Binding Motif
| Accessions: | ETV2_DLX3_2_3 (HumanTF2 1.0) |
| Names: | ETV2_DLX3 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 18 |
| Consensus: | rcCGGAAryrrgyaATTa |
| Weblogo: | 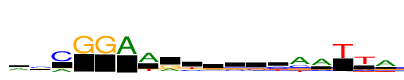 |
| PSSM: | P0 A C G T 01 3136 659 1636 824 r 02 1567 3195 1576 483 c 03 1622 4772 140 8 C 04 0 1 4772 6 G 05 4 0 4772 7 G 06 4772 0 0 24 A 07 4772 312 23 1435 A 08 1824 511 2354 83 r 09 706 1508 579 1978 y 10 1373 1023 1458 917 r 11 1472 1163 1528 608 r 12 942 1113 2008 709 g 13 680 2757 302 2015 y 14 4772 2066 1373 1284 a 15 4772 1211 612 406 A 16 175 108 125 4772 T 17 297 1226 464 4772 T 18 4772 218 1612 1005 a |
| Type: | Heterodimer |
| Binding TFs: | DLX3_TF2 (Homeobox domain) DLX3_Crystal (Homeobox domain) DLX3_Full (Homeobox domain, Homeobox protein distal-less-like N terminal ) ETV2_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.