DNA Binding Motif
| Accessions: | MA2001.1 (JASPAR 2024) |
| Names: | Six4 |
| Organisms: | Mus musculus |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Length: | 11 |
| Consensus: | gwAACCTGAbm |
| Weblogo: | 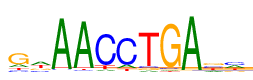 |
| PSSM: | P0 A C G T 01 701 471 2545 574 g 02 1495 629 779 1388 w 03 4063 99 57 72 A 04 3970 112 62 147 A 05 209 3506 115 461 C 06 480 3352 114 345 C 07 202 268 210 3611 T 08 124 146 3906 115 G 09 4124 62 52 53 A 10 835 1112 1239 1105 b 11 1344 1402 686 859 m |
| Type: | Heterodimer |
| Binding TFs: | Q61321 (Homeobox domain, Homeobox KN domain) Q61321 |
| Binding Sites: | MA2001.1.1 / MA2001.1.2 MA2001.1.10 / MA2001.1.14 MA2001.1.11 / MA2001.1.15 MA2001.1.12 / MA2001.1.17 MA2001.1.13 / MA2001.1.18 MA2001.1.14 / MA2001.1.19 MA2001.1.15 / MA2001.1.20 MA2001.1.16 MA2001.1.17 MA2001.1.18 MA2001.1.19 MA2001.1.2 / MA2001.1.3 MA2001.1.20 MA2001.1.3 / MA2001.1.4 MA2001.1.4 / MA2001.1.7 MA2001.1.5 / MA2001.1.8 MA2001.1.6 / MA2001.1.9 MA2001.1.11 / MA2001.1.7 MA2001.1.12 / MA2001.1.8 MA2001.1.13 / MA2001.1.9 MA2001.1.1 MA2001.1.10 MA2001.1.16 MA2001.1.5 MA2001.1.6 |
| Publications: | Himeda CL, Ranish JA, Angello JC, Maire P, Aebersold R, Hauschka SD. Quantitative proteomic identification of six4 as the trex-binding factor in the muscle creatine kinase enhancer. Mol Cell Biol 24:2132-43 (2004). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.