DNA Binding Motif
| Accessions: | ETV2_HES7_2_3 (HumanTF2 1.0) |
| Names: | ETV2_HES7 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 22 |
| Consensus: | rsCGGAwrygrhggCrCGyGsc |
| Weblogo: | 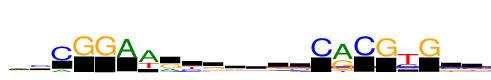 |
| PSSM: | P0 A C G T 01 2283 565 1609 719 r 02 842 2611 1281 293 s 03 770 3892 17 6 C 04 0 17 3892 0 G 05 13 0 3892 4 G 06 3892 10 28 66 A 07 3892 119 0 1805 w 08 1318 345 2191 37 r 09 274 1484 378 1756 y 10 635 651 2038 567 g 11 1275 830 1024 763 r 12 979 1036 888 988 h 13 913 516 2004 459 g 14 863 878 1814 337 g 15 63 3892 0 21 C 16 3892 42 2678 436 r 17 11 3892 0 15 C 18 26 20 3892 33 G 19 247 1757 95 3892 y 20 30 13 3892 52 G 21 197 1746 1087 862 s 22 446 1846 687 912 c |
| Type: | Heterodimer |
| Binding TFs: | HES7_TF2 (Helix-loop-helix DNA-binding domain) ETV2_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.