DNA Binding Motif
| Accessions: | ETV2_NHLH1_2 (HumanTF2 1.0) |
| Names: | ETV2_NHLH1 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 21 |
| Consensus: | wcmGGAwgkramGCAGCTGCk |
| Weblogo: | 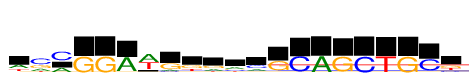 |
| PSSM: | P0 A C G T 01 2234 241 449 1324 w 02 1933 4247 2299 974 c 03 2162 4247 57 5 m 04 3 1 4247 8 G 05 0 8 4247 0 G 06 4247 175 13 56 A 07 4247 425 15 3633 w 08 802 612 2831 2 g 09 321 374 2001 1552 k 10 1441 212 1851 744 r 11 1731 826 887 803 a 12 1371 2033 473 370 m 13 181 151 3559 357 G 14 19 4247 0 49 C 15 4247 10 0 0 A 16 27 69 4247 22 G 17 25 4247 0 0 C 18 25 0 4 4247 T 19 8 0 4247 18 G 20 169 3806 112 159 C 21 92 930 1713 1513 k |
| Type: | Heterodimer |
| Binding TFs: | NHLH1_TF2 (Helix-loop-helix DNA-binding domain) ETV2_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.