DNA Binding Motif
| Accessions: | ETV2_HES7_1 (HumanTF2 1.0) |
| Names: | ETV2_HES7 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 20 |
| Consensus: | ggCrCGyGsckrcCGGAwry |
| Weblogo: | 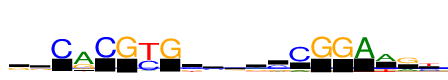 |
| PSSM: | P0 A C G T 01 887 648 2763 781 g 02 1098 1141 2306 533 g 03 112 5079 10 41 C 04 5079 179 2178 411 r 05 11 5079 45 17 C 06 30 0 5079 19 G 07 296 2922 183 5079 y 08 58 0 5079 64 G 09 170 2102 1584 1223 s 10 950 2130 1139 860 c 11 1005 918 1830 1327 k 12 2099 391 1805 784 r 13 738 2971 1202 169 c 14 840 5079 115 36 C 15 6 12 5079 29 G 16 2 80 5079 13 G 17 5079 7 16 56 A 18 3519 273 0 1560 w 19 1591 340 3489 121 r 20 349 2228 474 2851 y |
| Type: | Heterodimer |
| Binding TFs: | HES7_TF2 (Helix-loop-helix DNA-binding domain) ETV2_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.