DNA Binding Motif
| Accessions: | IRX3_2 (HT-SELEX2 May2017), UN0130.1 (JASPAR 2024) |
| Names: | IRX3 |
| Organisms: | Homo sapiens |
| Libraries: | HT-SELEX2 May2017 1, JASPAR 2024 2 1 Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schübeler D, Vinson C, Taipale J. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science : (2017). [Pubmed] 2 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | HT-SELEX |
| Length: | 18 |
| Consensus: | wACAyGawwwwtCrTGTw |
| Weblogo: | 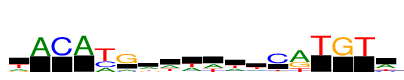 |
| PSSM: | P0 A C G T 01 3636 2 2284 6765 w 02 12686 73 0 34 A 03 18 12686 1 154 C 04 12686 0 2 4 A 05 4569 6163 28 12686 y 06 2319 1454 12686 306 G 07 6019 2038 1858 2773 a 08 5806 791 882 5207 w 09 5789 472 461 5964 w 10 6173 427 481 5606 w 11 5255 916 845 5670 w 12 2958 1832 2056 5840 t 13 370 12686 1342 2021 C 14 12686 24 6050 4491 r 15 0 2 0 12686 T 16 145 5 12686 22 G 17 32 0 74 12686 T 18 6804 2303 15 3566 w |
| Type: | Heterodimer |
| Binding TFs: | IRX3 (Homeobox domain, Homeobox KN domain) P78415 (Homeobox domain, Homeobox KN domain) P78415 |
| Publications: | Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schübeler D, Vinson C, Taipale J. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science : (2017). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.