DNA Binding Motif
| Accessions: | GCM1_FIGLA_2 (HumanTF2 1.0) |
| Names: | GCM1_FIGLA |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 23 |
| Consensus: | CAssTGkkrbkkkrksrTGcGGG |
| Weblogo: | 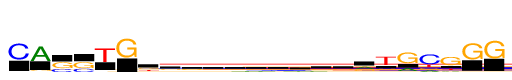 |
| PSSM: | P0 A C G T 01 21 1495 78 36 C 02 1495 64 78 46 A 03 16 544 951 41 s 04 26 564 930 15 s 05 87 55 85 1495 T 06 16 0 1495 22 G 07 102 298 639 456 k 08 279 285 505 426 k 09 384 325 501 284 r 10 254 428 434 378 b 11 345 267 477 405 k 12 277 343 496 379 k 13 282 337 487 388 k 14 387 313 450 344 r 15 304 274 531 385 k 16 234 414 502 344 s 17 697 174 535 88 r 18 131 157 341 1495 T 19 459 114 1495 10 G 20 311 1495 397 410 c 21 211 94 1495 355 G 22 16 45 1495 61 G 23 17 19 1495 32 G |
| Type: | Heterodimer |
| Binding TFs: | FIGLA_TF2 (Helix-loop-helix DNA-binding domain) GCM1_TF1 (GCM motif protein) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.