DNA Binding Motif
| Accessions: | ETV2_PITX1 (HumanTF2 1.0) |
| Names: | ETV2_PITX1 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 19 |
| Consensus: | rsCGGAArtrrrGGmTTAg |
| Weblogo: | 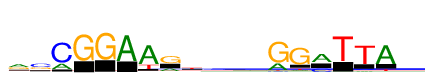 |
| PSSM: | P0 A C G T 01 905 108 701 218 r 02 246 992 614 90 s 03 456 1606 33 10 C 04 0 7 1606 3 G 05 0 0 1606 0 G 06 1606 0 5 18 A 07 1606 81 8 404 A 08 445 71 1065 24 r 09 179 379 271 777 t 10 449 361 544 252 r 11 582 255 501 268 r 12 491 300 516 299 r 13 532 80 1606 90 G 14 129 64 1606 166 G 15 1094 512 25 30 m 16 2 16 35 1606 T 17 95 119 47 1606 T 18 1606 29 56 199 A 19 380 399 573 253 g |
| Type: | Heterodimer |
| Binding TFs: | PITX1_TF1 / PITX1_TF2 (Homeobox domain) ETV2_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.