DNA Binding Motif
| Accessions: | ETV2_TEF_2 (HumanTF2 1.0) |
| Names: | ETV2_TEF |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 23 |
| Consensus: | TTACGTAAccmmgwACCGGAAry |
| Weblogo: | 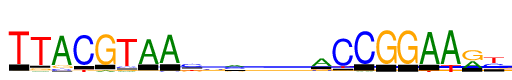 |
| PSSM: | P0 A C G T 01 0 10 33 1216 T 02 57 13 103 1216 T 03 1216 117 132 0 A 04 0 1216 22 114 C 05 80 13 1216 1 G 06 61 93 64 1216 T 07 1125 46 13 32 A 08 1155 52 3 5 A 09 67 676 204 269 c 10 230 548 300 138 c 11 489 487 206 35 m 12 333 512 181 190 m 13 221 297 487 211 g 14 322 240 258 396 w 15 845 124 158 89 A 16 43 1216 57 101 C 17 44 1216 0 0 C 18 0 20 1216 1 G 19 9 11 1216 0 G 20 1216 1 0 67 A 21 1216 4 17 271 A 22 489 3 725 0 r 23 0 499 102 616 y |
| Type: | Heterodimer |
| Binding TFs: | TEF_TF1 / TEF_TF2 (bZIP transcription factor, Basic region leucine zipper) ETV2_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.