DNA Binding Motif
| Accessions: | TFAP4_DLX3_2 (HumanTF2 1.0) |
| Names: | TFAP4_DLX3 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 21 |
| Consensus: | yCAGsTGkdbgkrsgtaATtr |
| Weblogo: | 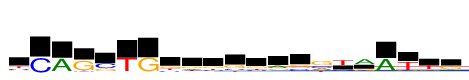 |
| PSSM: | P0 A C G T 01 145 267 168 354 y 02 0 933 12 0 C 03 933 44 3 32 A 04 0 99 933 114 G 05 66 597 337 16 s 06 0 36 16 933 T 07 25 0 933 0 G 08 74 215 341 304 k 09 244 129 322 238 d 10 135 234 316 248 b 11 44 222 490 179 g 12 145 217 337 234 k 13 431 124 251 127 r 14 59 336 456 82 s 15 462 482 933 366 g 16 112 220 152 933 t 17 933 98 337 89 a 18 933 18 33 26 A 19 70 37 166 933 T 20 84 138 256 677 t 21 325 85 608 121 r |
| Type: | Heterodimer |
| Binding TFs: | DLX3_TF2 (Homeobox domain) TFAP4_TF1 (Helix-loop-helix DNA-binding domain) TFAP4_Full (Helix-loop-helix DNA-binding domain) DLX3_Crystal (Homeobox domain) DLX3_Full (Homeobox domain, Homeobox protein distal-less-like N terminal ) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.