DNA Binding Motif
| Accessions: | GCM1_FIGLA_1 (HumanTF2 1.0) |
| Names: | GCM1_FIGLA |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 15 |
| Consensus: | CAssTGsaCCCgCAy |
| Weblogo: | 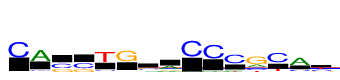 |
| PSSM: | P0 A C G T 01 30 1107 12 20 C 02 1107 125 56 48 A 03 19 629 478 47 s 04 28 667 440 30 s 05 41 165 76 1107 T 06 70 94 1107 66 G 07 182 289 428 208 s 08 623 112 260 112 a 09 13 1107 22 23 C 10 50 1107 47 24 C 11 164 1107 43 150 C 12 406 401 1107 224 g 13 12 1107 65 341 C 14 1107 254 96 96 A 15 80 514 207 593 y |
| Type: | Heterodimer |
| Binding TFs: | FIGLA_TF2 (Helix-loop-helix DNA-binding domain) GCM1_TF1 (GCM motif protein) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.