DNA Binding Motif
| Accessions: | TFAP2C_MAX_2_3_4 (HumanTF2 1.0) |
| Names: | TFAP2C_MAX |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 31 |
| Consensus: | tgsCCysrGGssayrysssvssrrsCACGTG |
| Weblogo: | 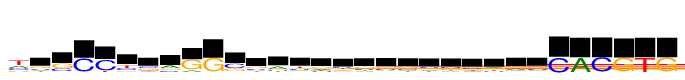 |
| PSSM: | P0 A C G T 01 6491 6257 4061 12025 t 02 2653 2747 4310 2315 g 03 328 4679 7346 522 s 04 65 12025 628 61 C 05 19 12025 438 1854 C 06 406 4240 2186 5192 y 07 1970 4269 3754 2032 s 08 5407 2600 3702 316 r 09 2407 336 12025 102 G 10 85 316 12025 48 G 11 481 7365 4660 453 s 12 2436 3380 3880 2328 s 13 5584 2146 2337 1957 a 14 2667 3419 1976 3962 y 15 3472 2319 3640 2593 r 16 2746 3441 2740 3098 y 17 2431 3386 3721 2487 s 18 2385 3483 4280 1878 s 19 2639 3165 3940 2281 s 20 3114 3438 3193 2279 v 21 2446 3712 3299 2569 s 22 2652 3324 3325 2725 s 23 3346 2370 3313 2995 r 24 4014 2003 4329 1678 r 25 2621 3457 3951 1996 s 26 94 12025 52 0 C 27 12025 89 85 97 A 28 0 12025 77 164 C 29 260 36 12025 69 G 30 6 135 150 12025 T 31 214 0 12025 80 G |
| Type: | Heterodimer |
| Binding TFs: | MAX_TF2 (Helix-loop-helix DNA-binding domain) TFAP2C_TF1 (Transcription factor AP-2) MAX_Full (Helix-loop-helix DNA-binding domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.