DNA Binding Motif
| Accessions: | TFAP2C_ETV7 (HumanTF2 1.0) |
| Names: | TFAP2C_ETV7 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 26 |
| Consensus: | sCYbsrGGssrcGGAAgyvytTCCks |
| Weblogo: | 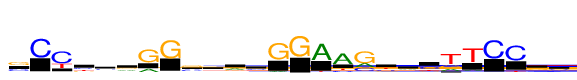 |
| PSSM: | P0 A C G T 01 33 154 226 22 s 02 0 380 5 7 C 03 3 380 17 148 Y 04 24 133 98 124 b 05 70 133 109 68 s 06 130 64 141 45 r 07 106 11 380 0 G 08 0 16 380 1 G 09 51 180 200 34 s 10 60 117 109 94 s 11 175 62 101 41 r 12 63 145 88 84 c 13 40 0 380 1 G 14 0 0 380 0 G 15 380 5 1 32 A 16 380 61 3 68 A 17 136 119 380 21 g 18 40 121 64 155 y 19 129 95 100 55 v 20 19 189 71 101 y 21 100 6 118 380 t 22 53 1 19 380 T 23 5 380 7 2 C 24 10 380 3 39 C 25 56 77 152 95 k 26 67 124 116 73 s |
| Type: | Heterodimer |
| Binding TFs: | TFAP2C_TF1 (Transcription factor AP-2) ETV7_TF1 / ETV7_TF2 (Ets-domain, Sterile alpha motif (SAM)/Pointed domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.