DNA Binding Motif
| Accessions: | SOX6_TBX21_2 (HumanTF2 1.0) |
| Names: | SOX6_TBX21 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 21 |
| Consensus: | rgGTGtkrwwdwdrACAATrg |
| Weblogo: | 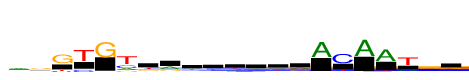 |
| PSSM: | P0 A C G T 01 544 108 305 233 r 02 284 219 565 150 g 03 8 131 849 164 G 04 8 205 22 849 T 05 28 4 849 20 G 06 274 330 188 849 t 07 98 163 269 319 k 08 431 116 213 89 r 09 231 184 208 227 w 10 234 189 200 226 w 11 214 150 236 250 d 12 254 178 198 218 w 13 252 140 241 217 d 14 353 123 229 144 r 15 849 2 28 30 A 16 12 849 89 162 C 17 849 6 5 24 A 18 849 32 37 89 A 19 216 69 66 849 T 20 355 122 494 192 r 21 153 193 342 162 g |
| Type: | Heterodimer |
| Binding TFs: | TBX21_TF2 (T-box) SOX6_TF1 / SOX6_TF2 (HMG (high mobility group) box, Domain of unknown function (DUF1898)) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.