DNA Binding Motif
| Accessions: | ETV2_SOX15_2_3 (HumanTF2 1.0) |
| Names: | ETV2_SOX15 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 20 |
| Consensus: | rCCGGAWGyamrmACAAwrg |
| Weblogo: | 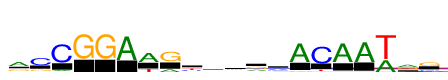 |
| PSSM: | P0 A C G T 01 816 114 406 94 r 02 100 961 261 65 C 03 114 1222 36 9 C 04 4 0 1222 0 G 05 3 1 1222 0 G 06 1222 12 8 0 A 07 885 36 0 337 W 08 292 37 907 0 G 09 135 385 132 570 y 10 465 257 275 226 a 11 455 387 129 251 m 12 405 120 608 89 r 13 572 312 239 98 m 14 1222 26 49 53 A 15 17 1222 50 140 C 16 1222 47 33 44 A 17 1222 19 27 53 A 18 617 13 2 1222 w 19 647 175 575 69 r 20 137 234 749 102 g |
| Type: | Heterodimer |
| Binding TFs: | SOX15_TF2 (HMG (high mobility group) box, Domain of unknown function (DUF1898)) ETV2_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.