DNA Binding Motif
| Accessions: | THRB_methyl_6 (HT-SELEX2 May2017) |
| Names: | THRB |
| Organisms: | Homo sapiens |
| Libraries: | HT-SELEX2 May2017 1 1 Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schübeler D, Vinson C, Taipale J. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science : (2017). [Pubmed] |
| Length: | 19 |
| Consensus: | rTGWCCTyRCrTGWCCTyR |
| Weblogo: | 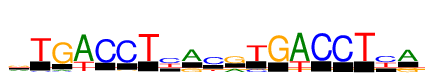 |
| PSSM: | P0 A C G T 01 240 24 263 118 r 02 0 30 4 615 T 03 54 33 645 0 G 04 645 0 0 238 W 05 5 645 19 16 C 06 8 645 32 17 C 07 1 0 9 645 T 08 0 369 60 236 y 09 475 0 168 5 R 10 34 458 43 111 C 11 207 20 406 11 r 12 0 111 0 533 T 13 0 0 645 16 G 14 645 0 0 254 W 15 0 645 4 0 C 16 3 645 3 5 C 17 0 0 0 645 T 18 0 380 37 264 y 19 465 0 201 0 R |
| Binding TFs: | THRB (Zinc finger, C4 type (two domains)) |
| Publications: | Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schübeler D, Vinson C, Taipale J. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science : (2017). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.