DNA Binding Motif
| Accessions: | UP00399A_1 (UniPROBE 20160601) |
| Names: | "POU domain, "POU2F1, class 2, NF-A1, Oct-1, OCT1, Octamer-binding transcription factor 1, OTF-1", OTF1, transcription factor 1" |
| Organisms: | Homo sapiens |
| Libraries: | UniPROBE 20160601 1 1 Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res : (2015). [Pubmed] |
| Length: | 22 |
| Consensus: | ywwradwTwTGCAwAwtsarwm |
| Weblogo: | 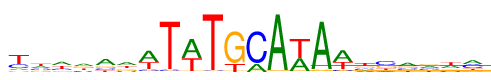 |
| PSSM: | P0 A C G T 01 0.11 0.25 0.10 0.53 y 02 0.27 0.21 0.21 0.31 w 03 0.34 0.18 0.15 0.33 w 04 0.31 0.19 0.26 0.24 r 05 0.48 0.17 0.19 0.16 a 06 0.34 0.15 0.25 0.26 d 07 0.54 0.09 0.04 0.33 w 08 0.02 0.04 0.01 0.93 T 09 0.58 0.02 0 0.40 w 10 0.01 0.02 0 0.97 T 11 0.01 0 0.78 0.21 G 12 0.21 0.78 0 0.01 C 13 0.97 0 0.02 0.01 A 14 0.40 0 0.02 0.58 w 15 0.93 0.01 0.04 0.02 A 16 0.55 0.08 0.04 0.34 w 17 0.20 0.21 0.23 0.36 t 18 0.14 0.40 0.28 0.18 s 19 0.37 0.22 0.24 0.17 a 20 0.28 0.23 0.30 0.19 r 21 0.31 0.15 0.22 0.33 w 22 0.36 0.35 0.14 0.15 m |
| Binding TFs: | UP00399A (Homeobox domain, Pou domain - N-terminal to homeobox domain) |
| Binding Sites: | ATGCAAAT ATATTCAT AATATGCA AATATTCA AATTATGC AATTATTC ACTAATTA ATATGCAA ATATGCAT ATGAATAA ATGCATAA ATTAATTA ATTATGCA ATTATTCA ATTTAAAT CTAATTAA GCTAATTA TATGAATA TATGCAAA TATGCATA |
| Publications: | Berger M.F, Philippakis A.A, Qureshi A.M, He F.S, Estep P.W, Bulyk M.L. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nature biotechnology 24:1429-35 (2006). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.