DNA Binding Motif
| Accessions: | TFAP2C_DLX3_2 (HumanTF2 1.0) |
| Names: | TFAP2C_DLX3 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 23 |
| Consensus: | gsCCysrRGscaygskrTAATkr |
| Weblogo: | 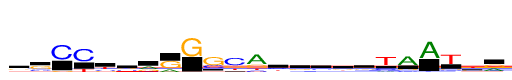 |
| PSSM: | P0 A C G T 01 408 433 639 392 g 02 186 671 1201 129 s 03 22 1872 225 13 C 04 8 1872 17 533 C 05 111 517 392 852 y 06 257 546 644 425 s 07 914 308 573 77 r 08 530 72 1342 5 R 09 18 3 1872 14 G 10 99 727 1872 207 s 11 135 1872 617 472 c 12 1872 362 762 397 a 13 307 715 322 528 y 14 355 346 747 424 g 15 445 619 471 336 s 16 423 358 566 524 k 17 490 378 639 364 r 18 264 529 82 1872 T 19 1872 266 303 96 A 20 1872 6 37 82 A 21 141 206 227 1872 T 22 266 310 761 1111 k 23 817 94 1055 105 r |
| Type: | Heterodimer |
| Binding TFs: | DLX3_TF2 (Homeobox domain) TFAP2C_TF1 (Transcription factor AP-2) DLX3_Crystal (Homeobox domain) DLX3_Full (Homeobox domain, Homeobox protein distal-less-like N terminal ) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.