DNA Binding Motif
| Accessions: | GCM2_HOXA13 (HumanTF2 1.0), UN0553.1 (JASPAR 2024) |
| Names: | GCM2_HOXA13, GCM2::HOXA13 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1, JASPAR 2024 2 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] 2 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX, HT-SELEX |
| Length: | 15 |
| Consensus: | ATRCGGGTaATAAAa |
| Weblogo: | 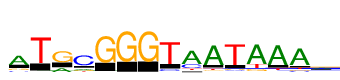 |
| PSSM: | P0 A C G T 01 5476 695 1108 191 A 02 215 122 72 7121 T 03 1924 92 5208 39 R 04 973 5940 798 653 C 05 157 37 7104 60 G 06 0 0 7191 0 G 07 0 0 7186 0 G 08 84 782 140 6984 T 09 6107 1975 190 837 a 10 7063 356 471 629 A 11 279 366 584 7103 T 12 6742 345 953 1602 A 13 7045 297 356 835 A 14 6817 624 648 726 A 15 3604 1656 989 936 a |
| Type: | Heterodimer |
| Binding TFs: | GCM2_TF1 (GCM motif protein) HOXA13_TF2 / P31271 (Homeobox domain, Hox protein A13 N terminal) HOXA13_Full (Homeobox domain, Hox protein A13 N terminal) P31271 |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.