DNA Binding Motif
| Accessions: | POU2F1_SOX15_2_3 (HumanTF2 1.0) |
| Names: | POU2F1_SOX15 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 16 |
| Consensus: | ywtGmATAACAAwrra |
| Weblogo: | 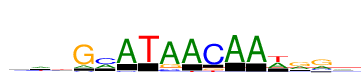 |
| PSSM: | P0 A C G T 01 2002 3722 1279 3521 y 02 4454 1963 593 3514 w 03 2343 2182 1408 4590 t 04 1539 577 10524 274 G 05 4104 6421 176 682 m 06 10524 674 16 221 A 07 172 46 202 10524 T 08 10524 1169 2232 430 A 09 10524 213 9 710 A 10 44 10524 1070 1021 C 11 10524 22 72 211 A 12 10524 289 47 105 A 13 5018 288 109 5506 w 14 4317 717 6207 1022 r 15 3690 2059 6835 173 r 16 4873 1939 2153 1559 a |
| Type: | Heterodimer |
| Binding TFs: | POU2F1_TF1 (Homeobox domain, Pou domain - N-terminal to homeobox domain) SOX15_TF2 (HMG (high mobility group) box, Domain of unknown function (DUF1898)) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.