DNA Binding Motif
| Accessions: | TFAP2C_MAX_2_3 (HumanTF2 1.0) |
| Names: | TFAP2C_MAX |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 31 |
| Consensus: | tgsCCysrGGssawrbsssssrrsCACGTGs |
| Weblogo: | 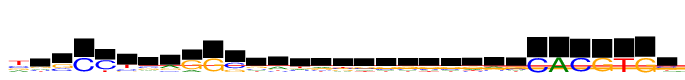 |
| PSSM: | P0 A C G T 01 5901 7469 6108 13804 t 02 3340 3200 4741 2522 g 03 538 5471 8333 788 s 04 0 13804 595 31 C 05 58 13804 571 3201 C 06 390 3958 2153 7302 y 07 2359 4716 4561 2167 s 08 6562 2477 4183 581 r 09 3673 388 13804 25 G 10 178 624 13804 20 G 11 259 9219 4584 578 s 12 2948 3534 4412 2909 s 13 6211 2378 2687 2527 a 14 3589 3337 2276 4601 w 15 4042 3004 3972 2785 r 16 2858 3641 3520 3784 b 17 3112 3681 3922 3088 s 18 3116 3758 4070 2860 s 19 2977 3570 4394 2863 s 20 2905 3899 4099 2901 s 21 3079 3819 4023 2882 s 22 3986 2622 4239 2956 r 23 4902 2605 4197 2099 r 24 2410 4179 4838 2377 s 25 128 13804 33 12 C 26 13804 86 16 8 A 27 21 13804 60 462 C 28 335 105 13804 36 G 29 73 222 186 13804 T 30 6 26 13804 112 G 31 1668 4928 4509 2698 s |
| Type: | Heterodimer |
| Binding TFs: | MAX_TF2 (Helix-loop-helix DNA-binding domain) TFAP2C_TF1 (Transcription factor AP-2) MAX_Full (Helix-loop-helix DNA-binding domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.