DNA Binding Motif
| Accessions: | MEIS1_DLX2 (HumanTF2 1.0) |
| Names: | MEIS1_DLX2 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 14 |
| Consensus: | tGACArswTAATtr |
| Weblogo: | 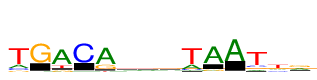 |
| PSSM: | P0 A C G T 01 2540 1745 1216 10866 t 02 415 195 10866 477 G 03 10866 204 1508 2199 A 04 494 10866 180 391 C 05 10866 138 3280 786 A 06 3452 1704 3854 1857 r 07 1720 3628 2915 2602 s 08 3432 1931 1932 3571 w 09 3203 1203 236 10866 T 10 10866 438 1328 275 A 11 10866 144 244 97 A 12 1356 1612 1452 10866 T 13 2192 2140 3193 7673 t 14 5345 1111 5521 1353 r |
| Type: | Heterodimer |
| Binding TFs: | DLX2_TF2 (Homeobox domain, Homeobox protein distal-less-like N terminal ) MEIS1_TF1 (Homeobox domain, Homeobox KN domain) DLX2_Full (Homeobox domain, Homeobox protein distal-less-like N terminal ) MEIS1_Crystal (Homeobox domain, Homeobox KN domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.