DNA Binding Motif
| Accessions: | MA0681.2 (JASPAR 2024) |
| Names: | PHOX2B |
| Organisms: | Homo sapiens, Mus musculus |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | ChIP-seq |
| Length: | 16 |
| Consensus: | wwTAATCAAATTAwww |
| Weblogo: | 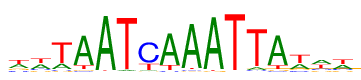 |
| PSSM: | P0 A C G T 01 18530 7301 8172 19233 w 02 19443 7036 6744 20013 w 03 10344 2748 2542 37602 T 04 42696 1935 2289 6316 A 05 51239 496 508 993 A 06 1742 615 467 50412 T 07 2451 36619 1873 12293 C 08 43074 1895 588 7679 A 09 48550 1453 1418 1815 A 10 50675 451 761 1349 A 11 921 329 372 51614 T 12 4103 1923 1232 45978 T 13 42869 1936 2155 6276 A 14 16088 4909 5569 26670 w 15 20366 8280 7630 16960 w 16 18565 7409 6999 20263 w |
| Type: | Heterodimer |
| Binding TFs: | Q99453 (Homeobox domain) Q99453 |
| Binding Sites: | MA0681.2.1 MA0681.2.10 MA0681.2.11 / MA0681.2.3 MA0681.2.12 MA0681.2.13 MA0681.2.14 MA0681.2.15 / MA0681.2.4 MA0681.2.16 / MA0681.2.5 MA0681.2.17 / MA0681.2.6 MA0681.2.18 MA0681.2.19 MA0681.2.2 MA0681.2.20 / MA0681.2.7 MA0681.2.3 MA0681.2.4 MA0681.2.5 MA0681.2.6 MA0681.2.1 / MA0681.2.7 MA0681.2.8 MA0681.2.2 / MA0681.2.9 MA0681.2.10 MA0681.2.11 MA0681.2.12 MA0681.2.13 MA0681.2.14 MA0681.2.15 MA0681.2.16 / MA0681.2.17 MA0681.2.18 MA0681.2.19 MA0681.2.20 MA0681.2.8 MA0681.2.9 |
| Publications: | Vincentz JW, VanDusen NJ, Fleming AB, Rubart M, Firulli BA, Howard MJ, Firulli AB. A Phox2- and Hand2-dependent Hand1 cis-regulatory element reveals a unique gene dosage requirement for Hand2 during sympathetic neurogenesis. J Neurosci 32:2110-20 (2012). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.