DNA Binding Motif
| Accessions: | ERF_DLX2_2 (HumanTF2 1.0) |
| Names: | ERF_DLX2 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 18 |
| Consensus: | rsmGGAwrtsrvymaTTa |
| Weblogo: | 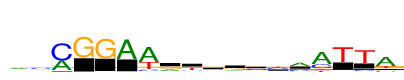 |
| PSSM: | P0 A C G T 01 474 147 257 148 r 02 169 421 309 105 s 03 418 730 24 0 m 04 2 0 730 8 G 05 2 2 730 19 G 06 730 17 11 0 A 07 730 95 0 357 w 08 294 73 311 53 r 09 116 147 81 387 t 10 166 186 263 116 s 11 210 150 315 55 r 12 200 240 216 75 v 13 110 357 67 374 y 14 477 253 94 41 m 15 730 253 122 110 a 16 25 32 50 730 T 17 50 118 11 730 T 18 730 91 97 176 a |
| Type: | Heterodimer |
| Binding TFs: | DLX2_TF2 (Homeobox domain, Homeobox protein distal-less-like N terminal ) DLX2_Full (Homeobox domain, Homeobox protein distal-less-like N terminal ) ERF_TF1 (Ets-domain) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.