DNA Binding Motif
| Accessions: | GCM1_FIGLA_2_3_4 (HumanTF2 1.0) |
| Names: | GCM1_FIGLA |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 20 |
| Consensus: | CAssTGkdrbgkgrTGcGGG |
| Weblogo: | 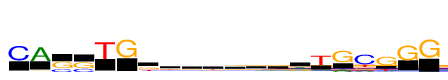 |
| PSSM: | P0 A C G T 01 55 1693 82 64 C 02 1693 26 130 95 A 03 18 548 1144 46 s 04 45 635 1058 12 s 05 23 61 25 1693 T 06 35 13 1693 17 G 07 120 265 731 577 k 08 468 277 441 507 d 09 425 324 613 331 r 10 240 426 505 522 b 11 409 375 527 382 g 12 374 323 562 434 k 13 257 417 621 398 g 14 747 233 601 112 r 15 146 180 214 1693 T 16 430 60 1693 24 G 17 339 1693 444 516 c 18 199 91 1693 422 G 19 46 44 1693 93 G 20 30 17 1693 14 G |
| Type: | Heterodimer |
| Binding TFs: | FIGLA_TF2 (Helix-loop-helix DNA-binding domain) GCM1_TF1 (GCM motif protein) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.